Protoplast fusion
and genetic analysis
in Cephalosporium acremonium
3.3.2 Screening of commercial enzymes for the
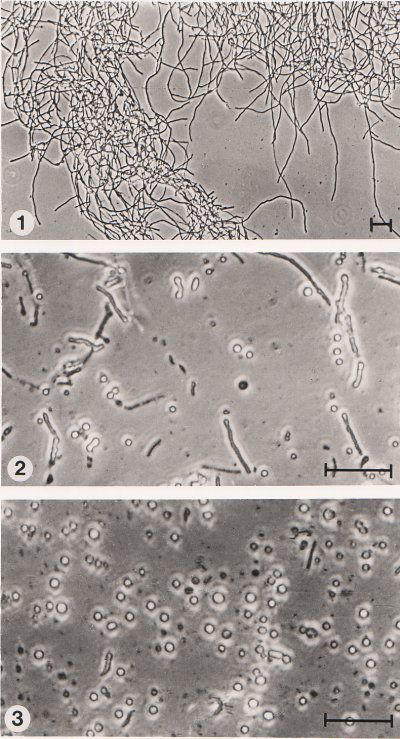
ability to induce protoplast Although high protoplast yields could be obtained with the laboratory preparation from Cytophaga it would be much more convenient to use a commercial product. Several enzymes, i.e. cellulase (Merck), chitinase (Sigma) and helicase which had been used previously for the isolation of protoplasts from fungi (AnnÚ, 1977; AnnÚ et al., 1974) were tried as well as other commercial polysaccharases normally used in the brewing and food industries. It seemed possible that the latter products could contain side activities capable of digesting fungal cell walls. The effectiveness of these enzymes in releasing protoplasts from C. acremonium is indicated in Table 3.3. Chitinase, ▀-D-glucanase, Cereflo 200L, Celluclast 200L and Finizym 200L individually failed to promote protoplast release and did not appear to have any lytic activity against Cephalosporium cell walls. Conversely, the other preparations were capable of producing protoplasts and the yields were enhanced in all cases following pretreatment of the mycelium with dithiothreitol. Cellulase (Merck) when used alone was not very effective but in combination with chitinase gave a high protoplast yield. However, combinations of enzymes which individually gave good protoplast yields (e.g. Novozym 234 + Cellulase CP) had no enhancing effect. It was apparent from this study that two of the enzyme preparations were particularly suitable for further investigations. Both Cellulase CP and Novozym 234 gave satisfactory protoplast yields on their own. Although these enzymes are not manufactured on a large scale they are relatively inexpensive and could prove useful for the routine production of fungal protoplasts. Observations on the release of protoplasts from filamentous fungi show that very often they emerge through pores in the hyphae leaving behind empty walls. In C. acremonium protoplasts were released from the hyphal strands after 10-15 min of lytic treatment. After 2-3 h of incubation the process was almost complete. Partially digested mycelial fragments could be removed from the lytic mixture by slow centrifugation to give a relatively clean preparation of protoplasts (Fig. 3.9).
3.3.7 Isolation of protoplasts from other species of fungi It was considered important to be able to carry out fusion experiments between closely related or more distant species of fungi without having to screen for new lytic enzymes on each occasion. Therefore, the most suitable enzyme would have to be active against a wide range of fungi. The lytic preparations described earlier have been tested against several other species including Aspergillus nidulans, Aspergillus niger, Penicillium chrysogenum, Volvariella volvacea and Saccharomyces cerevisiae by colleagues at Nottingham (Hamlyn et al., 1981). Under the conditions employed the most versatile preparation appeared to be Novozym 234 although with the Aspergilli a combination of Novozym 234 and Cellulase CP proved to be more effective (Table 3.6). However, Cellulase CP has now been shown to be more active at a lower pH and the effectiveness of this enzyme with other fungi needs to be reassessed.
References AnnÚ, J. (1977). Somatic hybridization between Penicillium species after induced fusion of their protoplasts. Agricultura, 25, 1-117. AnnÚ, J., Eyssen, H. and De Somer, P. (1974). Formation and regeneration of Penicillium chrysogenum protoplasts. Archives of Microbiology, 98, 159-166. Hamlyn, P.F., Bradshaw, R.E., Mellon, F.M., Santiago, C.M., Wilson, J.M. & Peberdy, J.F. (1981). Efficient protoplast isolation from fungi using commercial enzymes. Enzyme Microbial Technology, 3, 321-325. |
| TABLE 3.3 Comparison of commercial enzymes for their ability to induce protoplast release from mycelium of C. acremonium |
| Enzyme | Protoplast yield (x106 ml-1)* | |
| Pretreatment with dithiothreitol |
No pretreatment |
|
| Cellulase (Merck) | 8.00 ▒ 3.27 | 0.75 ▒ 0.54 |
| Cellulase CP (Sturge) | 133 ▒ 18 | 13.3 ▒ 6.2 |
| Cellulase CT (Sturge) | 1.67 ▒ 1.03 | 0.42 ▒ 0.12 |
| Chitinase (Sigma) | <0.25 | <0.25 |
| Cellulase (Merck) + Chitinase (Sigma) | 85.0 ▒ 12.2 | Not tested |
| ▀-D-Glucanase (BDH) | <0.25 | <0.25 |
| Helicase (L'Industrie Biologique) | 93.3 ▒ 10.3 | 5.83 ▒ 3.12 |
| Lytic enzyme L1 (BDH) | 237 ▒ 29 | 81.7 ▒ 6.2 |
| Novozym 234 (Novo) | 148 ▒ 12 | 9.3 ▒ 2.49 |
| Cereflo 200L (Novo) | <0.25 | <0.25 |
| Celluclast 200L (Novo) | <0.25 | <0.25 |
| Finizym 200L (Novo) | <0.25 | <0.25 |
*The lytic mixture contained 5ml of 0.7M NaCl in buffer (pH 5.8), mycelium (250 mg; damp-dry) and the lytic enzyme (solid enzymes 25mg; liquid enzymes 5%). Lytic digestions were carried out in triplicate and the values quoted are means ▒ standard deviation. Note: Some of these enzymes may no longer be commercially available and the companies may have changed their name. For example, John & E Sturge Ltd (Selby, England) is now Tate & Lyle Citric Acid. |
| TABLE 3.6 Isolation of protoplasts from other species of fungi using the commercial enzymes1 |
| Enzyme |
Protoplast yield (x106 ml-1) |
Protoplast yield2 |
|||
| A. nidulans | A. niger | P. chrysogenum | V. volvacea | S. cerevisiae | |
| Cellulase CP | 19 ▒ 5 | 0.94 ▒ 0.O6 | 8.3 | <0.25 | 36.4 ▒ 21.9 |
| Novozym 234 | 190 ▒ 53 | 1.57 ▒ 0.7 | 37.9 ▒ 5 | 1.53 ▒ 0.05 | 98.9 ▒ 0.3 |
| Cellulase CP + Novozym 234 |
280 ▒ 14 | 17.9 ▒ 0.3 |
| 1After Hamlyn et al., (1981). Results are expressed as
mean ▒ standard deviation. 2With S. cerevisiae protoplast yield = % cells as protoplasts.
|

Figure 3.9 Formation of protoplasts from mycelium of C. acremonium after treatment with lytic enzyme. (1) Mycelium before lytic digestion. (2) After 1 h of incubation with the enzyme showing fragmented mycelium and protoplasts. (3) Suspension of protoplasts after 3 h followed by slow centrifugation to remove the mycelial debris. The bar markers represent 30 Ám. |
Copyright ę 1982 Paul F Hamlyn
(http://fungus.org.uk/cv/thesis_fig3.9.htm)